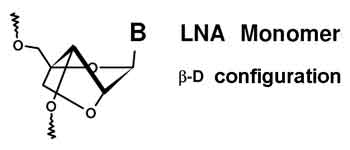
A locked nucleic acid (LNA), often referred to as inaccessible RNA, is a modified RNA nucleotide. The ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form of DNA or RNA. LNA nucleotides can be mixed with DNA or RNA bases in the oligonucleotide whenever desired. Such oligomers are commercially available. The locked ribose conformation enhances base stacking and backbone pre-organization. This significantly increases the thermal stability (melting temperature) of oligonucleotides.[1]
LNA was independently invented by the laboratories of Jesper Wengel[2] and Takeshi Imanishi[3] in 1998.
LNA nucleotides are used to increases the sensitivity and specificity of expression in DNA microarrays, FISH probes, real-time PCR probes and other molecular biology techniques based on oligonucleotides. For the in situ detection of miRNA the use of LNA is currently (2005) the only efficient method. A triplet of LNA nucleotides surrounding a single-base mismatch site maximizes LNA probe specificity unless the probe contains the guanine base of G-T mismatch.[4]
A new hepatitis C drug based on a LNA targeting a microRNA is in clinical testing as of late 2009.[5]
Benefits of the LNA technology
Some of the benefits of using LNA include:
* Ideal for the detection of short RNA and DNA targets
* Increases the thermal stability of duplexes
* Capable of single nucleotide discrimination
* Resistant to exo- and endonucleases resulting in high stability in vivo and in vitro applications
* Increased target specificity
* Facilitates Tm normalization
* Strand invasion properties enables detection of “hard to access” samples
* Compatible with standard enzymatic processes[6]
References
1. ^ Kaur, H; Arora, A; Wengel, J; Maiti, S; Arora, A.; Wengel, J.; Maiti, S. (2006). "Thermodynamic, Counterion, and Hydration Effects for the Incorporation of Locked Nucleic Acid Nucleotides into DNA Duplexes". Biochemistry 45 (23): 7347–55. doi:10.1021/bi060307w. PMID 16752924.
2. ^ Alexei A. Koshkin; Sanjay K. Singh, Poul Nielsen, Vivek K. Rajwanshi, Ravindra Kumar, Michael Meldgaard, Carl Erik Olsen, Jesper Wengel (1998). "LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition". Tetrahedron 54 (14): 3607–30. doi:10.1016/S0040-4020(98)00094-5.
3. ^ Satoshi Obika; Daishu Nanbu, Yoshiyuki Hari, Jun-ichi Andoh, Ken-ichiro Morio, Takefumi Doi, Takeshi Imanishi (1998). "Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2'-O,4'-C-methyleneribonucleosides". Tetrahedron Lett. 39 (30): 5401–4. doi:10.1016/S0040-4039(98)01084-3.
4. ^ You Y.; Moreira B.G.; Behlke M.A. and Owczarzy R. (2006). "Design of LNA probes that improve mismatch discrimination". Nucleic Acids Res. 34 (8): e60. doi:10.1093/nar/gkl175. PMID 16670427. PMC 1456327. http://nar.oxfordjournals.org/cgi/reprint/34/8/e60?ijkey=IqLeijzpwQHCzit&keytype=ref.
5. ^ Emily Singer. (2009). "Hepatitis C Drug Targets RNA". Technology Review. http://www.technologyreview.com/biomedicine/24059/.
6. ^ Locked Nucleic Acid Technology
External links
* LNA Oligo Tools and Design Guidelines
* LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA
* LNA: a versatile tool for therapeutics and genomics
* Alternative Nucleic Acid Analogues for Programmable Assembly: Hybridization of LNA to PNA
Retrieved from "http://en.wikipedia.org/"
All text is available under the terms of the GNU Free Documentation License