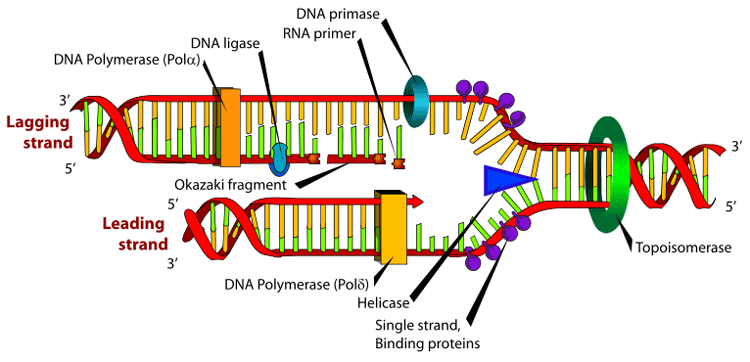
Many enzymes are involved in the DNA replication fork. An Okazaki fragment is a relatively short fragment of DNA (with no RNA primer at the 5' terminus) created on the lagging strand during DNA replication. The lengths of Okazaki fragments are between 1,000 to 2,000 nucleotides long in E. coli and are generally between 100 to 200 nucleotides long in eukaryotes. It was originally discovered in 1968 by Reiji Okazaki, Tsuneko Okazaki, and their colleagues while studying replication of bacteriophage DNA in Escherichia coli.[1][2] When the lagging strand is being replicated on the original strand, the 5'-3' pattern must be used; thus a small discontinuity occurs and an Okazaki fragment forms. These fragments are processed by the replication machinery to produce a continuous strand of DNA and hence a complete daughter DNA helix. In dealing with the synthesis of complementary DNA strands the nascent leading strand always reads 3' to 5'. Its antiparallel complement strand, the nascent lagging strand reads from 5' to 3'. Because the original strands of DNA are antiparallel, and only one continuous new strand can be synthesised at the 3' end of the leading strand due to the intrinsic 5'-3' polarity of DNA polymerases, the other strand must grow discontinuously in the opposite direction. Regarding the lagging strand, the result of this strand's discontinuous replication is the production of a series of short sections of DNA called Okazaki fragments. Each Okazaki fragment is initiated near the replication fork at an RNA primer created by primase, and extended by DNA polymerase III. In eukaryotes, lagging strand synthesis is carried out by the DNA polymerase δ. The primer is later removed by enzymes that have endonucleolytic activity such as Ribonuclease H (RNAse H), flap endonucleases (FENs) and Dna2 helicase/nucleases. In prokaryotes the FEN nuclease is a domain of DNA polymerase I while in eukaryotes FENs are separate enzymes. The excised RNA bases are replaced with DNA by DNA polymerase I in prokaryotes or DNA polymerase δ in eukaryotes. Adjoining fragments are then linked together by DNA ligase, using phosphodiester bonds, to create a continuous strand of DNA. Okazaki used a pulse chase type experiment to confirm discontinuous strand replication. He took actively replicating DNA, then added "hot" tritiated nucleotides for a short pulse of about 5 seconds. During the 5 seconds the radioactive nucleotides were incorporated into the growing DNA strands. After the pulse Okazaki chased with "cold" un-labeled nucleotides for varying amounts of time and quickly isolated the DNA. Then the DNA was centrifuged and analyzed for radioactivity. What Okazaki found was that with short chases of about 7 to 15 seconds most of the radioactivity was found in the small fragments higher in the tube after centrifuge. However with longer chases more radioactivity was found in the lower, larger strands. This confirmed that during synthesis first small fragments are formed on the lagging strand, then later these fragments are combined and incorporated into much larger strands. The small fragments found on the lagging strand are called Okazaki fragments. References 1. ^ Okazaki R, Okazaki T, Sakabe K, Sugimoto K. Mechanism of DNA replication possible discontinuity of DNA chain growth. Jpn J Med Sci Biol. 1967 Jun;20(3):255-60. Notes * Inman RB, Schnos M, Structure of branch points in replicating DNA: Presence of single-stranded connections in lambda DNA branch points. J. Mol Biol. 56:319-625, 1971.
* MeSH Okazaki+fragments Retrieved from "http://en.wikipedia.org/"
|