Piwi-interacting RNA (piRNA) is the largest class of small RNA molecules that is expressed in animal cells[1][2]. piRNA forms RNA-protein complexes through interactions with Piwi proteins. These piRNA complexes have been linked to transcriptional gene silencing of retrotransposons and other genetic elements in germ line cells, particularly those in spermatogenesis. They are distinct from miRNA in size (26–31 nt rather than 21–24 nt), lack of primary sequence conservation, and increased complexity [1][2]. It remains unclear how piRNAs are generated, but potential methods have been suggested, and it is certain their biogenesis pathway is distinct from miRNA and siRNA, while rasiRNAs are a piRNA subspecies[3].
Proposed piRNA structure piRNAs have been identified in both vertebrates and invertebrates, and although biogenesis and modes of action do vary somewhat between species, a number of features are conserved. piRNAs have no clear secondary structure motifs [1],.[4] the length of a piRNA is, by definition, between 26 and 31 nucleotides, and the presence of a 5’ uridine is common to piRNAs in both vertebrates and invertebrates. piRNAs in C. elegans have a 5’ monophosphate and a 3’ modification that acts to block either the 2’ or 3’ oxygen [5], and this has also been confirmed to exist in D.melanogaster[6], zebrafish [7], mice [8] and rats [7]. This 3’ modification is likely to be a 2’-O-methylation, but the reason for this modification is not known [7][9]. It is thought that there are many hundreds of thousands of different piRNA species found in mammals[10]. Thus far, over 50,000 unique piRNA sequences have been discovered in mice and more than 13,000 in D.melanogaster[11]. Location piRNAs are found in clusters throughout the genome; these clusters may contain as few as ten or up to many thousands of piRNAs[1][12] and can vary in size from one to one hundred kb [12]. While the clustering of piRNAs is highly conserved across species, the sequences are not[1][13]. While D.melanogaster and vertebrate piRNAs have been located in areas lacking any protein coding genes[3][14], piRNAs in C. elegans have been identified amidst protein coding genes [5]. In mammals, piRNAs are found only within the testes[7], with an estimated one million copies per cell in spermatocytes and spermatids[15]. In invertebrates, piRNAs have been detected in both the male and female germlines, but in no other cell types[7][10]. At the cellular level, piRNAs have been found within both nuclei and cytoplasm, suggesting that piRNA pathways may function in both of these areas[3] and, therefore, may have multiple effects[16]. Biogenesis
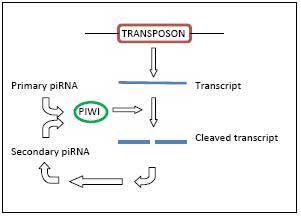
piRNA ping pong mechanism The biogenesis of piRNAs is not yet fully understood, although possible mechanisms have been proposed. piRNAs show a significant strand bias, that is, they are derived from one strand of DNA only[1], and this may indicate that they are the product of long single stranded precursor molecules[2]. A primary processing pathway is suggested to be the only pathway used to produce pachytene piRNAs; in this mechanism, piRNA precurors are transcribed resulting in piRNAs with a tendency to target 5’ uridines[17][18]. Also proposed is a ‘Ping Pong’ mechanism wherein primary piRNAs recognise their complementary targets and cause the recruitment of Piwi proteins. This results in the cleavage of the transcript at a point ten nucleotides from the 5’ end of the primary piRNA, producing the secondary piRNA[18]. These secondary piRNAs are targeted toward sequences that possess an adenine at the tenth position[17]. Since the piRNA involved in the ping pong cycle directs its attacks on transposon transcripts, the ping pong cycle acts only at the level of transcription[13]. One or both of these mechanisms may be acting in different species; C. elegans, for instance, does have piRNAs, but does not appear to use the ping pong mechanism at all[10]. A significant number of piRNAs identified in zebrafish and D.melanogaster contain adenine at their tenth position[3], and this has been interpreted as possible evidence of a conserved biosynthetic mechanism across species[9]. piRNAs are expressed through unique pathways, which are dissimilar to the expression pathways utilised by other small RNAs. Neither Argonaute proteins, RNA-dependent RNA polymerases, nor other proteins required for most small-RNA biosynthesis are necessary for piRNA expression[7][10]. However, while PIWI proteins are not required for the biosynthesis of piRNAs, evidence suggests that PIWI proteins must be present to stabilise piRNAs and, thus, facilitate their accumulation[10]. No mechanism for the control of piRNA propagation has yet been established[12]. Function The wide variation in piRNA sequences and PIWI function over species contributes to the difficulty in establishing the functionality of piRNAs[19]. However, like other small RNAs, piRNAs are thought to be involved in gene silencing[1], specifically the silencing of transposons. The majority of piRNAs are antisense to transposon sequences[13], suggesting that transposons are the piRNA target. In mammals it appears that the activity of piRNAs in transposon silencing is most important during the development of the embryo[17], and in both C. elegans and humans, piRNAs are necessary for spermatogenesis[19]. RNA Silencing piRNA has a role in RNA silencing via the formation of an RNA-induced silencing complex (RISC). piRNAs interact with Piwi proteins that are part of a family of proteins called the Argonautes. These are active in the testes of mammals and are required for germ-cell and stem-cell development in invertebrates. Three Piwi subfamily proteins - MIWI, MIWI2 and MILI - have been found to be essential for spermatogenesis in mice. piRNAs direct the Piwi proteins to their transposon targets[17]. A decrease or absence of PIWI protein expression is correlated with an increased expression of transposons[3][17]. Transposons have a high potential to cause deleterious effect on their host[12], and, in fact, mutations in piRNA pathways are found to reduce fertility in D.melanogaster[14]. However, piRNA pathway mutations in mice do not demonstrate reduced fertility; this may indicate redundancies to the piRNA system[3]. Further, it is thought that piRNA and endogenous small interfering RNA (endo-siRNA) may have comparable and even redundant functionality in transposon control in mammalian oocytes[13]. piRNAs appear to have an impact on particular methyltransferases that perform the methylations which are required to recognise and silence transposons[17], but this relationship is not well understood. Epigenetic Effects piRNAs can be transmitted maternally[7], and based on research in D.melanogaster, piRNAs may be involved in maternally derived epigenetic effects[14]. The activity of specific piRNAs in the epigenetic process also requires interactions between Piwi proteins and HP1a, as well as other factors[11]. Recent discovery also show, the existence of snoRNA, microRNA, piRNA characteristics in a novel non-coding RNA: x-ncRNA and its biological implication in Homo sapiens.[4] Investigation Due to their small size, expression and amplification of small RNAs can be challenging, so specialised PCR-based methods have been developed in response to this difficulty[20][21]. References 1. ^ a b c d e f g Molecular Biology Select. Cell, 2006. 126(2): p. 223, 225-223, 225.
* piRNA Bank A web resource on classified and clustered piRNAs
* N.C. Lau et al., "Characterization of the piRNA Complex from Rat Testes," Science 313, 363 (2006) Retrieved from "http://en.wikipedia.org/"
|